We will develop machine learning methods to classify protein-protein docking poses as correct or incorrect. We will improve the balance of the training set by employing SMOTE and GANs and the variance and size by the Snorkel technique. Our methods will be applicable to life sciences and bioengineering.
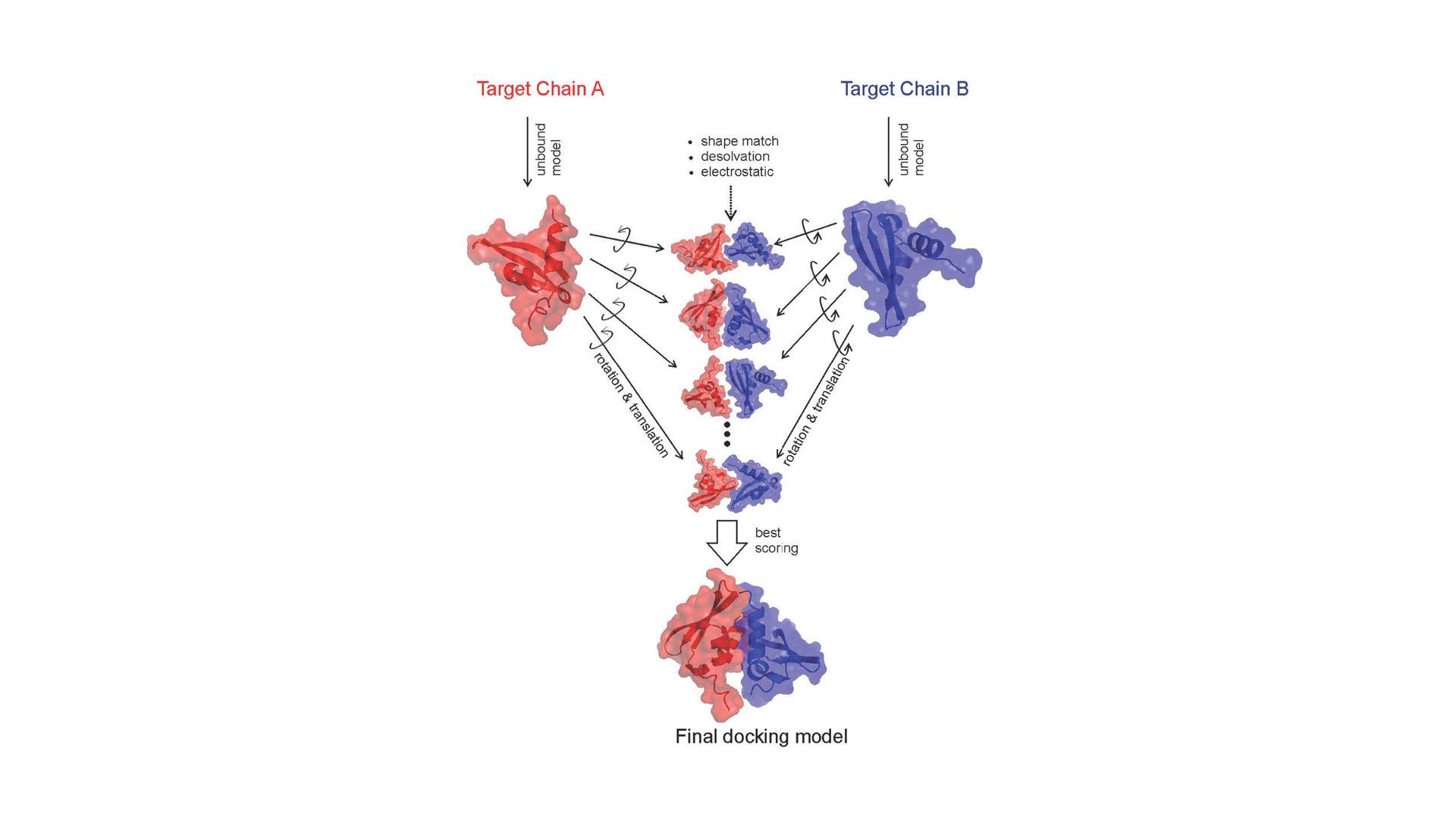
In this project we focus on protein-protein docking, a fundamental problem in computational biosciences with numerous applications, such as understanding Alzheimer’s disease, or designing enzymes for the biodegradation of pollutants in the environment. To date, reliably predicting the 3D structure of protein-protein complexes by molecular docking is still an open challenge, with one of the critical steps being the scoring, i.e. the ability to discriminate between correct and incorrect solutions within a wide pool of generated 3D poses. Motivated by this, we plan to utilize machine learning methods to classify protein-protein docking poses as correct or incorrect. We claim that classification accuracy can be improved significantly by improving the characteristics of the training data. We will improve the balance of the training set by employing SMOTE and GANs and the variance and size by the Snorkel technique. Our methods will be applicable to life sciences and bioengineering.
Investigator: