Polymers offer a better view
A window opens for analyzing the distribution of small molecules in biological and medical tissue samples.
About
Improvements in how samples are prepared will add range and flexibility to a method that detects the location of selected molecules within a biological sample, such as a slice of tissue.
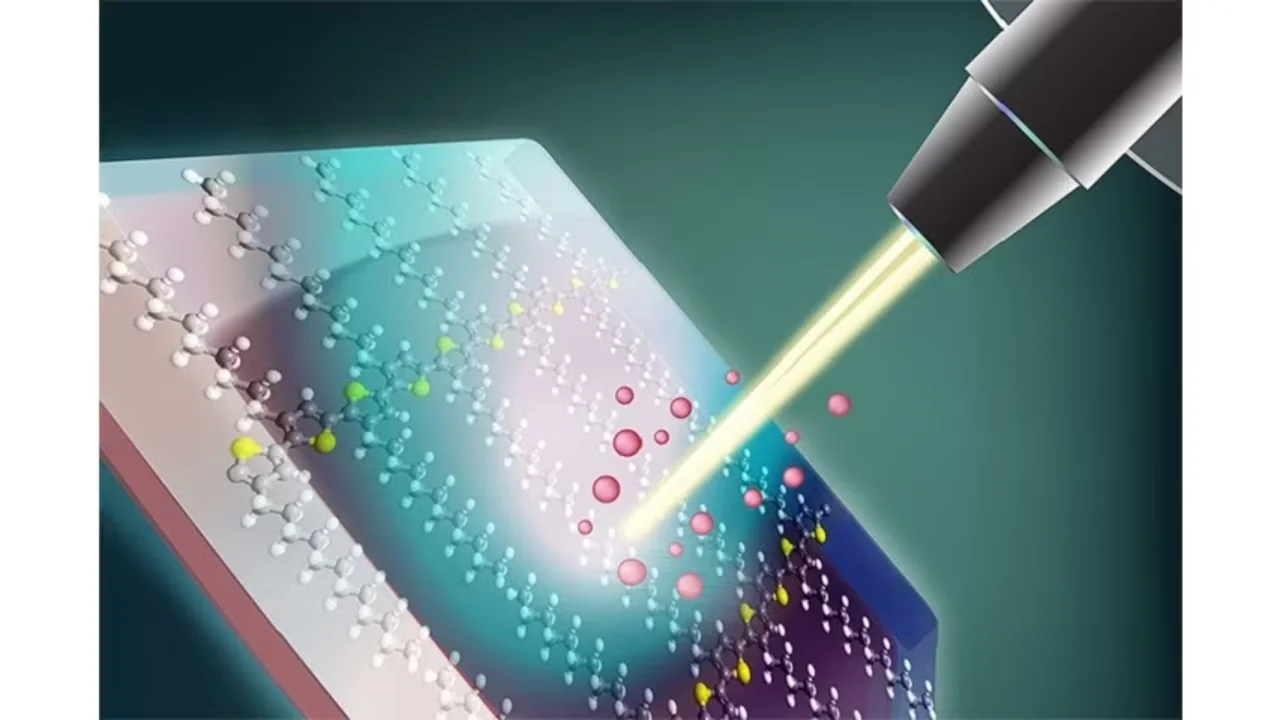
In the chemical analysis tool known as matrix-assisted laser-desorption/ionization mass spectrometry (MALDI MS), a laser beam kicks charged particles, known as ions, out of the sample. The ions are then fed through a mass spectrometer that detects them based on their mass. Repeating this process thousands of times, while the sample is moved in two dimensions, generates images that reveal the distribution of selected molecules throughout the sample. This process enables researchers to study the role of specific chemicals in biological and pathological applications.
Before the sample can be analyzed in this way, it must be embedded in a material called a matrix, but the small molecules generally used to form matrices impose limitations on the technique. Detecting metabolites, small molecules of biological and medical interest, has been particularly difficult due to interfering signals from molecules of the matrix.
Now a new class of matrices made of polymers that have larger molecules than conventional matrices has been developed by Dominik L. Michels from KAUST’s Visual Computing Center together with former KAUST postdoctoral researcher Franziska Lissel, who is now a junior faculty researching at the Leibniz Institute of Polymer Research (IPF) in Germany. In addition, the KAUST Catalysis Center's Nikos Hadjichristidis was essential to the success of the project.
“This eliminates many of the disadvantages of small molecule matrices,” says Lissel. She explains that the new polymer matrices enable the tracking of many more of the chemicals of interest in studying cancer as well as the location of drugs, which were previously unaccessible using MALDI imaging. “There are so many more research questions we can now explore,” she adds.
Read the full article
