Zooming in on brain function
Faster computations will allow researchers to see the finer details of brain activity in functional brain imaging.
About
A computationally efficient data-processing scheme will make it possible to see correlations in brain activity between different parts of the brain at unprecedented resolution. The statistics-based computation scheme, developed by KAUST researchers, also tackles one of the most critical problems of medical and biological imaging—how to process imaging data fast enough to realize the full exploratory power of the latest high-resolution imaging techniques.
The development of functional magnetic resonance imaging (fMRI) in the early 1990s was one of those true Eureka moments for brain research. Using existing noninvasive MRI technology, fMRI maps the distribution of blood oxygen in the brain, which is closely correlated with brain activity. With fMRI, it is possible to take snapshots of brain activity in response to specific stimuli, such as speech, responding to memory questions or visual scenes.
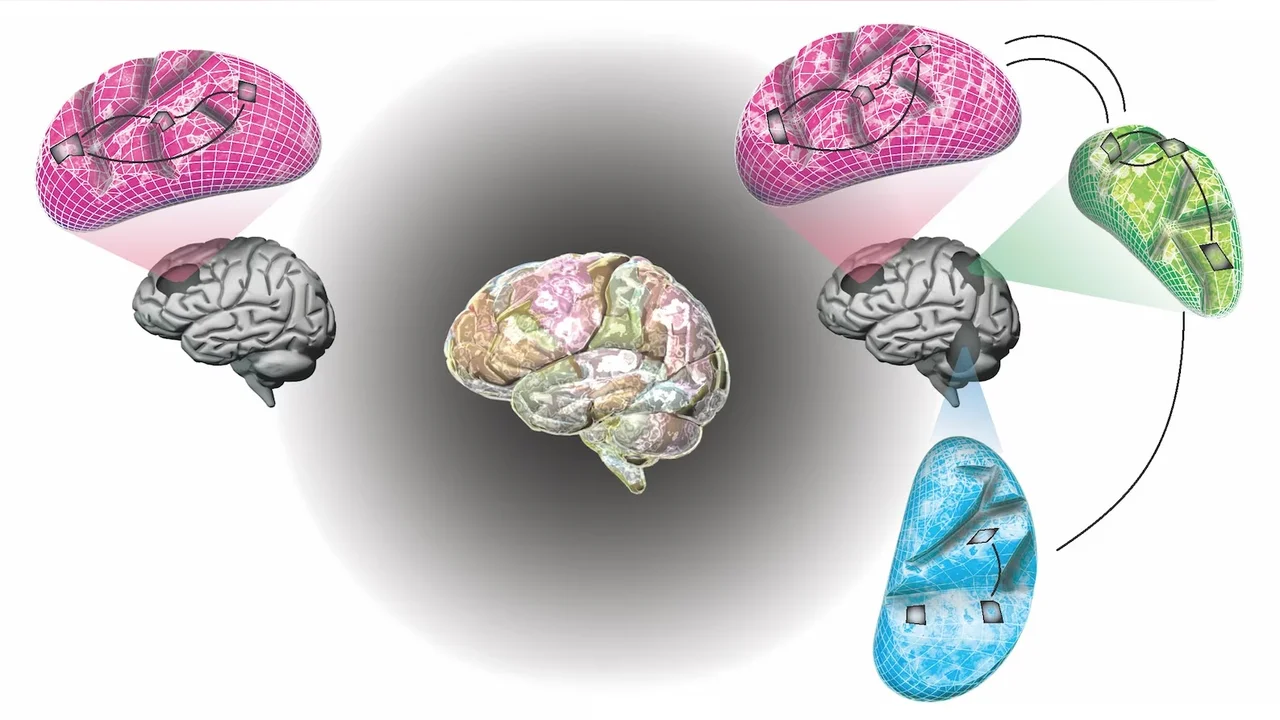
While fMRI is capable of taking high-resolution images consisting of hundreds of thousands of points or voxels, it is an enormous computational task to map the correlations between simultaneous brain function across different areas of the brain. Even with the range of computing power available today, such computations are not feasible by direct methods and so require a more computationally efficient approach.
Marc Genton and Hernando Ombao from KAUST, in collaboration with Stefano Castruccio from the University of Notre Dame in the United States, have addressed this problem. They developed a statistics-based computational scheme that matches activity in different parts of the brain at different spatial scales, from whole-of-brain to smaller regional structures and down to the tiniest brain volume.
“Using a statistical approach, our multiresolution approach essentially breaks up the spatial component of the fMRI data into different scales—from global to local,” says Ombao.
Read the full article

