© 2015 KAUST
Understanding the form and function of molecular complexes has important implications for the development of new drugs. Researchers from King Abdullah University of Science and Technology (KAUST) have devised a method to uncover detailed information about molecular complexes while overcoming the practical challenges of existing methods.
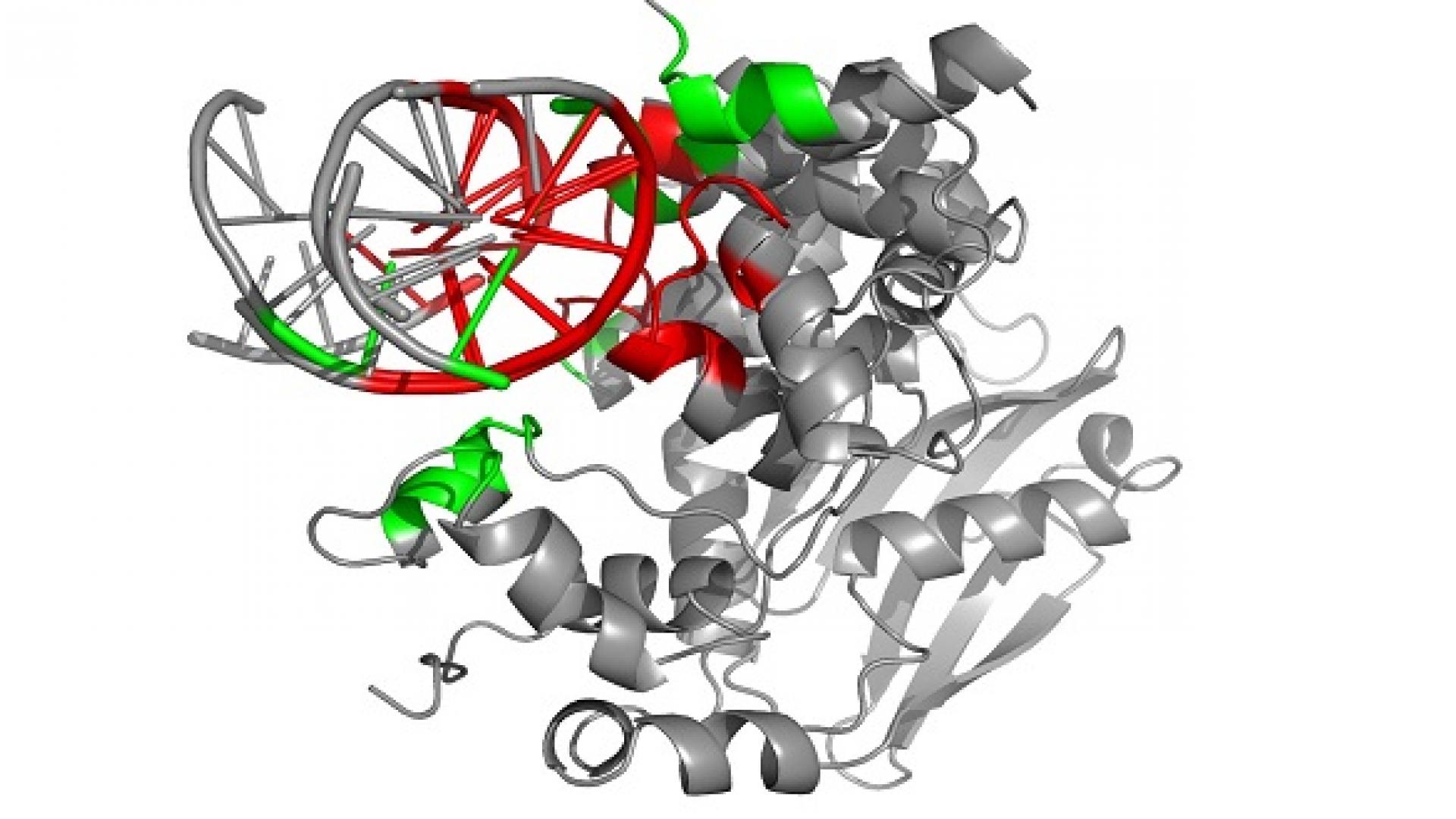
Biological molecules such as proteins, RNA and DNA perform their functions via interactions with other molecules, often forming connected "bundles" known as complexes. It is well-established that when these molecular complexes share a similar form, they are also likely to share a similar function.
Xin Gao, KAUST Assistant Professor of Computer Science, and colleagues from the University's Computer, Electrical and Mathematical Science and Engineering Division have developed a method called PROSTA-inter. The method can align the regions of individual molecules that connect together (known as the interaction interfaces) of two molecular complexes according to their 3D conformations.
Programs that can structurally align individual molecules allow scientists to match those that share similar functions. However, existing technology cannot align differing molecular complexes. For example, in aligning a protein-protein complex with a protein-RNA complex, existing techniques would overlook at least one of the interacting partners.
“Although the global structures of interacting pairs may be very different, they could have similar interaction interfaces and therefore similar functions,” explained Gao. “Existing methods cannot handle the alignment of complexes in full. For example, a model of a protein-DNA complex may be flawed because the program does not acknowledge the fact that the two DNA chains take turns to interact with the protein.”
Read the full article