Details
Technological breakthroughs in biological and biomedical research have led to the generation of a large amount of data in many areas. Utilizing this data for improving human life is nevertheless a challenge due to the inherent heterogeneity in the data, the complexity, and the large data size. The early promises of the success of next-generation sequencing in biomedicine have fallen short because it proved challenging to interpret the information obtained from individuals and translate it into new discoveries that benefit human health.
A key challenge in biomedicine is to understand the mechanisms underlying complex diseases in which multiple factors contribute, including environmental influences, and complex genetic and epigenetic influences. The most successful methods involve the integration of heterogeneous data from clinical and molecular studies, as well as from biological background knowledge. Similarly, identifying drug actions and their physiological effects crucially relies on understanding the physiology of a cell, interactions between multiple cells within a tissue and whole-body physiology, and ultimately requires the integration and analysis of multiple heterogeneous types of data.
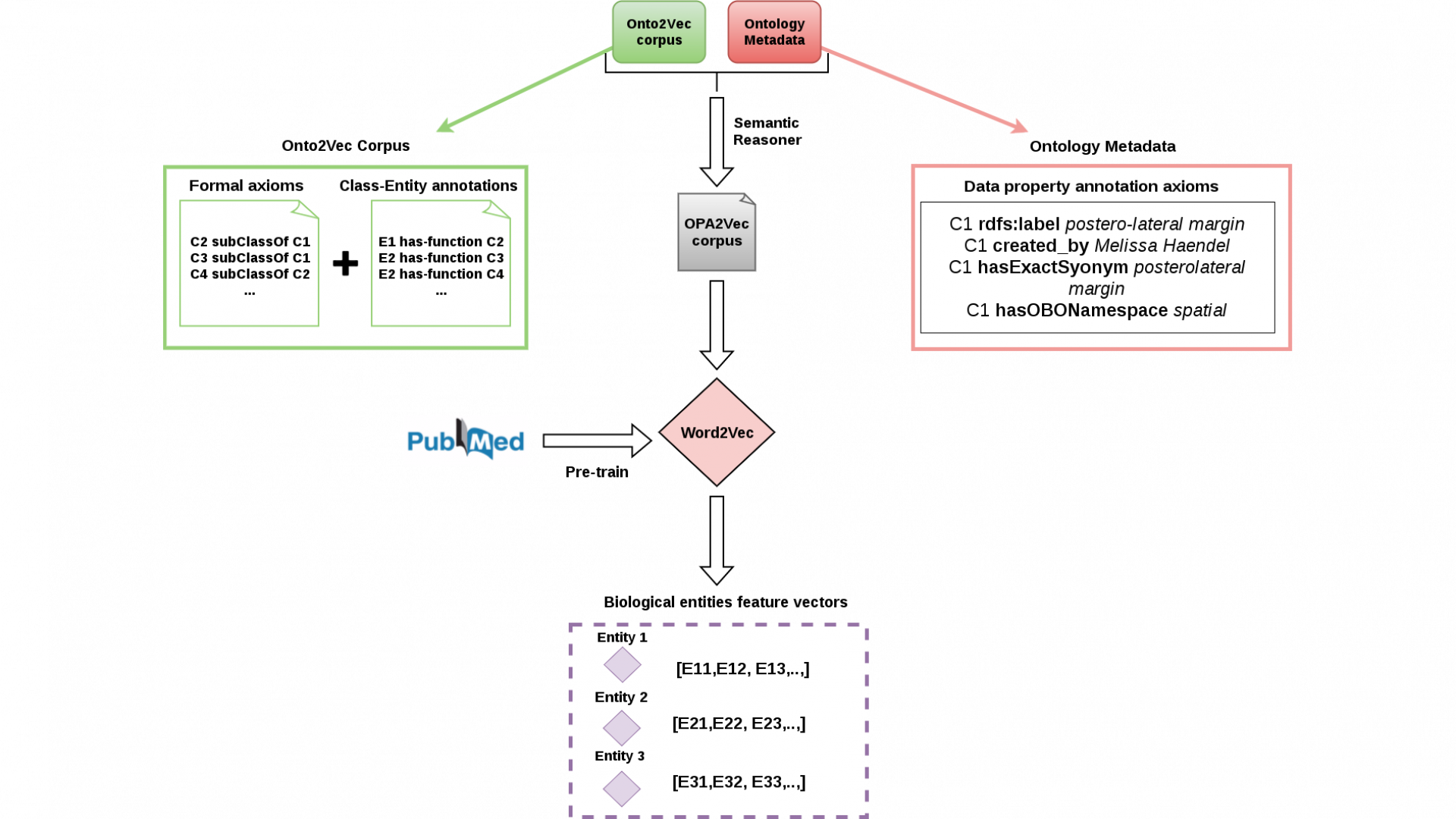
Several efforts aim to systematically provide information relevant to the interpretation of personal genomic data and interpretation of biological datasets in general. Many of these efforts are based on the use of knowledge graphs, i.e., information structured as heterogeneous graphs with explicit semantics, and are based on Semantic Web technologies. While these knowledge graphs have traditionally been used mainly to access information, several methods have recently become available that can utilize information in knowledge graphs directly for machine learning and data analysis.
However, these methods are not immediately applicable to biological and biomedical datasets due to the ways in which knowledge graphs in biology are constructed. Knowledge graphs in biology include a large number of biological and biomedical ontologies which provide rich background knowledge formalized in Description Logics that needs to be considered when using the information contained within them; biological knowledge graphs are of significant size (often containing billions of triples) and require tractable and scalable analytics methods. Furthermore, several biological data analysis methods have been developed for specific biological problems, such as different types of graph kernels, text mining methods that recognize specific biological entities, or sequence analysis methods, and these methods need to be utilized in conjunction with generic background knowledge contained in knowledge graphs to accurately interpret biological datasets.
In Bio2Vec, we will build the foundation for applying machine learning and data analytics methods on large scale to biological datasets. We specifically target the biological problems of computationally prioritizing disease modules underlying complex diseases, analysis of personal genomic information, and identifying physiological effects of drugs. The methods we will develop in Bio2Vec will be generically applicable throughout the life sciences and will improve data quality, representation, and analysis, and lead to novel discoveries in investigating molecular mechanisms underlying complex diseases and drugs’ mode of action.
Collaborators
- Michel Dumontier, Maastricht University
- Jens Lehmann, University of Bonn