Contact Person
Contact Person
Contact Person
Abstract
In July 2020, the National Center for Global Medicine (NCGM), in collaboration with SoftB
Contact Person
Abstract
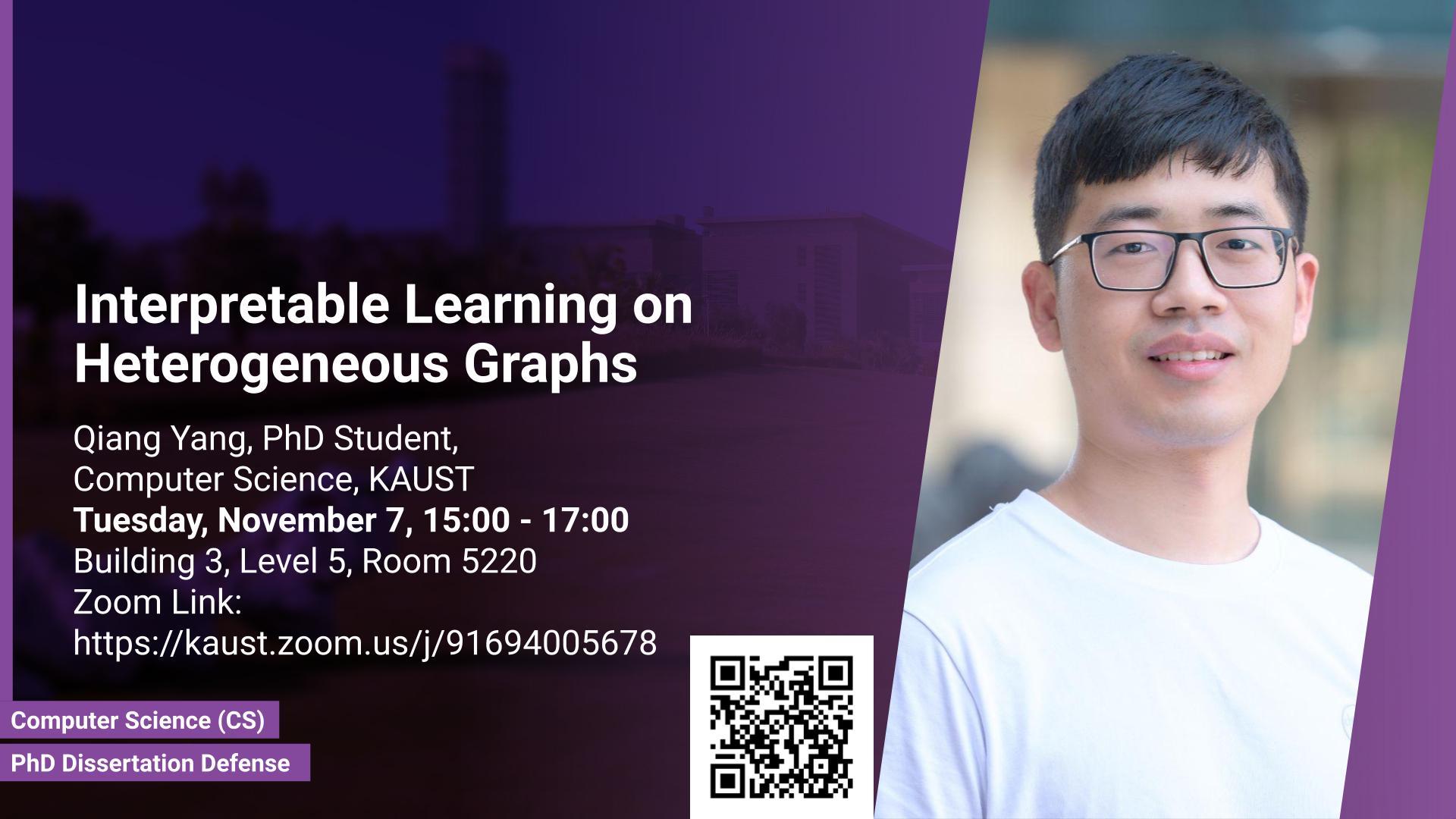
Graph regression is widely seen in material discovery as to predict numerical properties
Contact Person
Contact Person
Contact Person
KAUST - SFDA First Joint International Conference in Trends in Microbiome and Digital One Health
Contact Person
Abstract
Revoking personal private data is one of the basic human rights, which has already been s
Contact Person
This workshop aims to bring together distinguished and active scientists, from both academia and industry, i
Contact Person
Contact Person
Contact Person
Contact Person
Contact Person
The Computational Bioscience Research Center (CBRC) will be holding a student poster competition as part of
Contact Person
Computational Bioscience Research Center (CBRC) is pleased to announce the KAUST Research Conference 2023 on
Contact Person
Contact Person
Abstract
The increased availability of biomedical data, particularly in the public domain, offers
Contact Person
Abstract
The 2016 Global Burden of Disease Study identifies chronic kidney disease (CKD) as the 11
Contact Person
Contact Person
Abstract
In the coming years, expansion of crewed space exploration beyond low Earth orbit, to the
Contact Person
KAUST Research Open Week (KROW) is a campus-wide event with a range of activities taking place; from k