Analyses of two bacterial strains from the Red Sea show they are enriched with gene clusters with potential to activate the synthesis of a wide range of industrially useful compounds, from novel antibiotics, anticancer agents and pigments to those useful for crop protection and the food industry.
Bacteria are a rich resource for bioactive chemical compounds and Magbubah Essack, of KAUST’s Computational Bioscience Research Center, explains that bacterial strains able to withstand the Red Sea’s highly saline, warm waters were anticipated to produce sturdy enzymes suited for industrial applications.
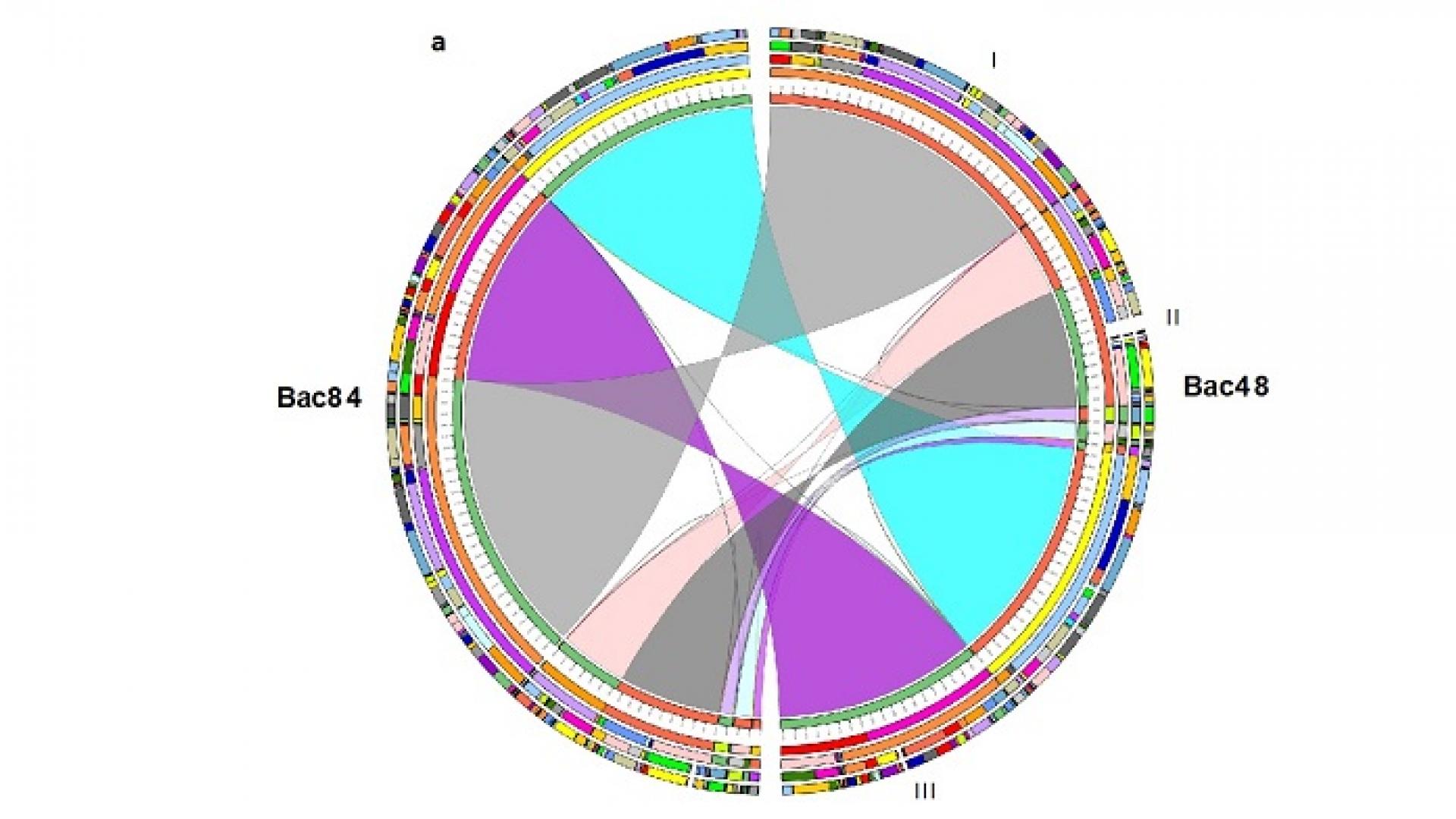
The team sequenced the genomes of two Bacillus species: B. paralicheniformis Bac48 collected from mangrove mud and B. paralicheniformis Bac84 collected from a microbial mat in the Rabigh Harbor Lagoon on Saudi Arabia’s west coast. These two were compared with the documented genomes of three other B. paralicheniformis and nine B. licheniformis strains. The Red Sea strains had a higher number of gene clusters associated with bioactive compound synthesis compared to the other Bacillus strains.

The research team report the complete circular and annotated genomes of two Red Sea strains, B. paralicheniformis Bac48 isolated from mangrove mud and B. paralicheniformis Bac84 isolated from microbial mat collected from Rabigh Harbor Lagoon in Saudi Arabia. (Front from l-r: Professor Takashi Gojobori, Professor Vladimir Bajic, Professor Heribert Hirt; Back from l-r: Dr. Magbubah Essack, Ameerah Bokhari, Dr. Salim Bougouffa, Associate Professor Xin Gao, Professor Stefan Arold.)
© 2018 KAUST
The team also report the first use of a computer program to identify a gene cluster in strains of the B. paralicheniformis species, in this case B. paralicheniformis Bac48, called trans-acyltransferase nonribosomal peptide synthetase/polyketide synthase, which is associated with the production of a specific group of compounds.
Read the full article